Experiment: Conformational analysis of butane
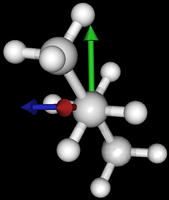
It's been quite a while since analysis_ethane, where I suggested that a look at butane might be interesting, too, as its structure of conformers is richer. As you can see by the following screenshots from MoleCuilder, there are four conformers.
| syn/eclipsed methyl | gauche | 120/eclipsed | anti |
 |  |  | 
|
However, the more complex an experiment, the more prone one is to making errors as well. I have made quite many: overwritten output files, stumbling over the limited precision of PDB files (use tremolo format if you're optimizing to very high precision), confusion when using the Python interface because of no "named arguments" support so far (contained in upcoming v1.6.1), copy&paste errors when building on the script I used for ethane.
But one thing especially impeded me: The quality of the optimization.
If one plans on investigating butane's rotational barriers, then one should have a fully annealed "anti" conformer state at hand.
But to make a long story short, here it is:
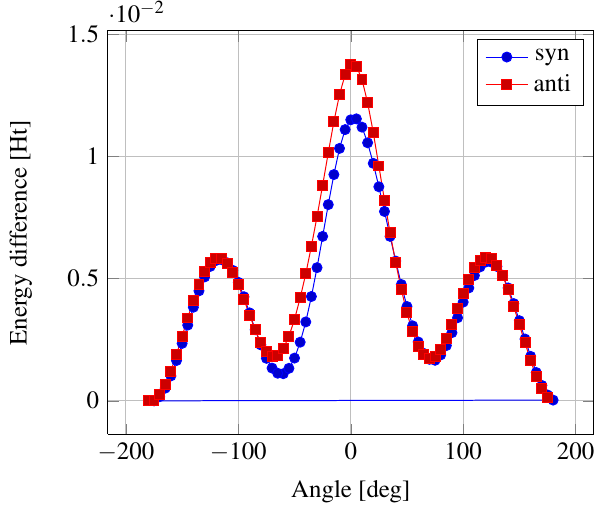
As you see in the conformer screenshots, I have done the rotation around the middle CC bond in the same manner as with ethane before. The conformer names relate to the following angles: syn (0°), gauche (60°), 120 (120°), anti (180°). I used two initial states: an optimized "syn" and an optimized "anti" state - both optimized to 1e-5 a.u. absolute error in energy in roughly 2000 steps. I have used 6-31G basis set and CLHF theory for optimization and rotation. If you'd like to reproduce the figure, using the docker image, both optimized conformer states are attached to this blog post including the update script to rotate around the CC bond and compute the energy. You need MoleCuilder version 1.6.1 which is soon to be released.
You notice that the two minima left and right of the central peak are not the same when starting from the "syn" state. They both correspond to the symmetric state of the same conformer. This is a clear indication that one needs to start from the lowest energy state, which is "anti". On the other hand, there is an energy difference in the central peak when starting from either state.
Comparing the above with experimental data at introorganicchemistry, you'll notice that the central peak is strongly overestimated by theory. However, a recent article by [Yirong Mo, JOCNote, 2012] states that experimental data and theoretical predictions do not match. This is why there are articles such as [Murcko, Castejon, Wiberg, 1996] and [Allinger, Fermann, Allen, Schaefer, JCP, 1997] calculating the energy differences to very high precision, estimating the syn/anti difference at around 0.087 Ht, while experimentally it is just 0.006 Ht, in chemical units 3.78 kcal/mole.
Here, we notice that the "syn" state is closer to the true energy difference as expected from theory, namely 0.0115 Ht. Note that [Yirong Mo, JOCNote, 2012] obtains roughly 0.01 Ht using Moeller-Plesset 2nd order and 6-31G(d) (with diffuse basis functions) where he optimized in each rotational step.
Having not optimized each step itself, I am satisfied with the results.
Using MoleCuilder in a docker container
Since the release of version v1.6.0, MoleCuilder is available as a debian package (installable at least under Ubuntu 14.04).
Since that it is pretty easy to build a docker image such that MoleCuilder can be run inside a docker container. A Dockerfile is provided in the same blog post.
To make it even easier, I created a public repository at Docker, where the image can be pulled from directly: frederikheber:molecuilder/latest
In other words and assuming you have a working docker installation at hand, all you need to do to try the latest version is the following.
docker pull frederikheber/molecuilder:latest docker run -it frederikheber/molecuilder:latest
This will open a root shell in the container. Type
molecuilder --version
And you'll see the version installed.
For computations you need both molecuilder_server and at least one instance of molecuilder_poolworker running. The Poolworker is the number cruncher. It's a modified version of MPQC. The Server accepts computation jobs from the MoleCuilder program and pushes them on to an idle worker. We start either one in the background, logging to a file:
molecuilder_server --controllerport 10024 --workerport 10025 2&>server.log & molecuilder_poolworker --server 127.0.0.1:10025 --listen 10026 --hostname 127.0.0.1 2&>worker.log &
The server listens to MoleCuilder on port 10024, the Worker enrolls with the Server on port 10025, and finally the Worker accepts jobs on 10026. Simply choose any ports available. You can also add more than one worker at a time using GNU's parallel, which is not installed on the container,
apt-get install parallel parallel --gnu molecuilder_poolworker --server 127.0.0.1:10025 --hostname 127.0.0.1 --listen ::: 10027 10028 &
Check the server.log file to see that indeed 3 workers are in the queue now.
Well, that's all. Would you like to calculate the energy of a methane molecule?
molecuilder \ --input methane.data \ --set-output tremolo \ --set-tremolo-atomdata "id type x=3 F=3 neighbors=4" --reset 1 \ --set-parser-parameters mpqc --parser-parameters "basis=STO-3G;theory=CLHF;" \ --add-atom C --domain-position "10,10,10" \ --select-atom-by-element C --saturate-atoms \ --select-all-atoms --optimize-structure --steps 10 --deltat 1. --server-address 127.0.0.1 --server-port 10024
This should bring the largest remaining force components to about 1e-5 a.u..
Don't be overwhelmed by the lot of typing and let's explain step by step:
- First, we specify an input file (which is empty but will contain the methane on exit).
- Next, we set the output format, namely tremolo.
- And afterwards, we need to specify the tremolo format to contain the id of the atom, its element, three spatial and three gradient coordinates and at most four (covalent) neighbor ids.
- The ab-initio calculations will be done at Closed-Shell Hartree Fock (CLHF) with a simple STO-3G basis set. As we use MPQC as number cruncher, we have to give these information to the parser mpqc.
- Then, finally, we create the methane molecule by adding a carbon atom and saturating it.
- Last, all atoms are selected and the structural optimization is performed for 10 steps with an initial step length of 1 a.u.. Notice that we give the port 10024, which we told the Server to use for listening to molecuilder's jobs.
Wanna take it from here? Increase the optimization steps. Choose another basis set, e.g. 6-31G (any of MPQC's sets is fine), or Moeller-Plesset Perturbation theory 2nd order (MBPT2), ...
Some pro tips:
- have one executable running per container, i.e. start you server as
docker run --name "MoleCuilderServer" -p 10024:10024 -p 10025:10025 frederikheber/molecuilder:latest \ /usr/bin/molecuilder_server --signal 2 15 --controllerport 10024 --workerport 10025
- similarly, start you workers each in a single container, too, where we automatically extract the servers IP address
SERVERIP=`docker inspect MolecuilderServer | grep IPAddress | tail -n 1 | awk -F":" '{print $2}' | tr -d \"\ ,` docker run --name "MoleCuilderWorker" -p 10026:10026 frederikheber/molecuilder:latest \ mpirun -np 1 /usr/bin/molecuilder_poolworker --signal 2 15 --server ${SERVERIP}:10025 --listen 10026 - use GNU's parallel to start e.g. 8 workers at once, assume the above is contained in script start_worker.sh
parallel --gnu --delay 1 --ctrlc ./start_worker.sh ::: `seq 1 8`
- you can also set up docker to start containers automatically at startup, see here. But this seems to be changing in the docker API.
- install molecuilder on you local machine and use molecuildergui for the methane optimization example above.

 rss
rss