Version 1.4.10 features tesselated surfaces as new molecule view in GUI, working List of Molecules, and some more.
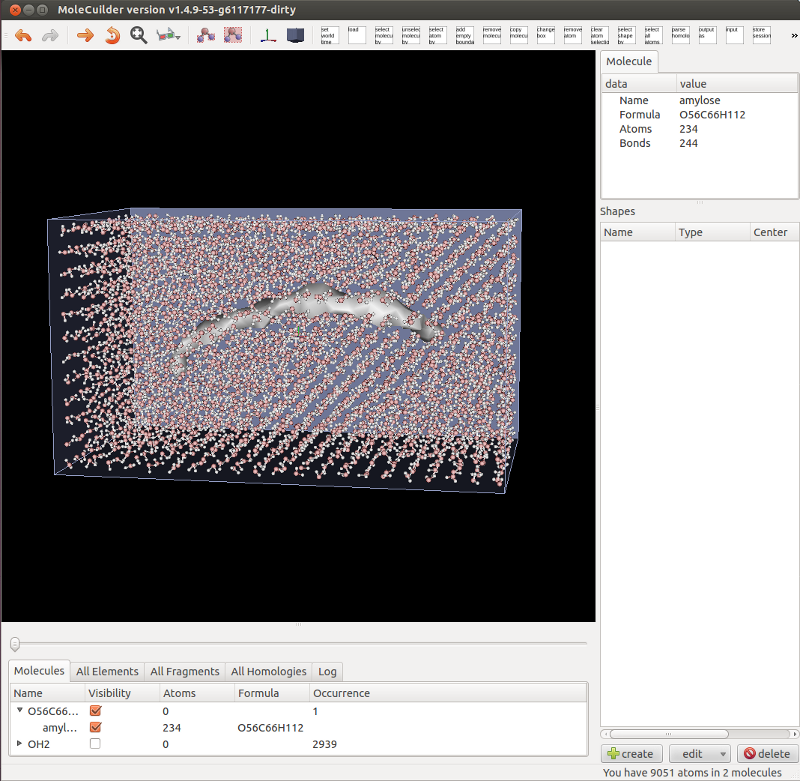
First of all, you can now simply flip a switch and molecules are no longer shown via their atoms and bonds but as a tesselated surface, see the image below of an amylose molecule filled with a regular grid of water molecules. This is especially useful with very large molecules (think e.g. a Tobacco Mosaic Virus or simply some large biomolecule) because a lot less triangles have to be drawn on the screen. Molecules can still be highlighted and selected by moving over or clicking onto the tesselated surface.
Notice also in the picture that the list of molecules is fully working (and hopefully) perfectly. Molecules are aggregated by their formula and can be individually selected or their visibility changed as well as for all of the same formula at once. Changing a molecule by adding an atom immediately changes the list. This rewrite became necessary as empty molecules are now automatically removed.
Furthermore, there's a number of smaller changes that should make the GUI use more easy going:
- the 3D view can now be changed via the keyboard (up/down, left/right, page-up/page-down and zooming with +/-)
- New RemoveMolecule action for convenience (before you had to select-molecules-atoms and remove-atoms)
- Selections can be pushed and popped which is useful inside MakroActions.
- Bonds are restored when undoing a remove action
- Failing actions no longer clean the queue and thus the destroy the stored session file
- when checking the command-line options for a specific action,
.../molecuilder --help fragment-molecule
is enough, actionname is no longer needed.

 rss
rss