Version v1.7.0 Makes Empirical Potentials First-Class Citizens
It's been 7 years since the last version was released: Now v1.7.0 is out! (see docker, source tarballs, and handbook)
I was working on walking the chemical space and subsequently on fixing so called "compound potentials" from ab-initio calculations. This necessitated some changes and also fixes which you find in a bit more detail below.
I plan to follow-up till the end of the year with a series of small posts where I highlight the more important changes. I want to end the series with a detailed explanation on how to fit your own set of empirical potentials for a given molecular system. Note that there are projects like https://openkim.org/ whose goal is to create a database of potential parameters that have been tested in deriving specific macroscopic quantities. I see the potentials more as a mean of saving computational effort: As the BOSSANOVA fragmentation approach is at the core of molecuilder, we end up running a lot of repetitive computations whose result we already know. In principle, the Born-Oppenheimer Surface (BOS) whose dynamics we compute can be very complex (narrow tunnels or deep potential holes). However, the empirical potentials keep it simple and we rely on the assumption that interactions are simple but their combined effects can be complex, i.e., it's the geometry that causes complexity.
List of Changes
Major
- saturate (fixes to polyhedra and scaling) and bondify (new)
- potentials are first class citizens
- proper path from molecule, to automated fragmentation, computation, homologies and finally potential training (and visualizing in the UI).
- convenience changes in the UI: bigger icons, context menu in gl view, potential tab with plotting
- enhancements to python scripting: numpy compatibility, wait returns status, python3
- works inside jupyter notebooks.
- can be deployed to k8s for large-scale computations.
- graph6 strings, evaluating stability and chemical space evaluation
- more selection actions
Minor
- switch to simpler kubernetes-aimed worker-server setup (health probes, workers don't register but accessed through load balancer)
- potential list shown in UI, new actions to add and remove, also potential curves are plotted in UI
- general fixes to potential setup to make them work as compound potentials
- undomark action
- context menu (right-click) in gl view to manipulate atoms directly
- docker file contained in repo
- bondify has undo/redo
- switched all python to python3
- graph6 fixes and writing strings
- generatePotentialAction generates all combinations
- full numpy getter and setter to atom position, velocities, and forces.
- saturate action allows for just using outer shell (unocc and occ orbitals, only unocc used for adding hydrogens)
- stretch bond falls back to tabled distances for element pairs on zero length
- select atom's bond neighbors
- select atoms randomly
- calculate pair correlation between element and selected atoms.
Fixes
- corrected units and computation in SaveEnergies, renamed from SaveTemperatures
- InputAction will also save that file on exit
Using MoleCuilder in a docker container
Since the release of version v1.6.0, MoleCuilder is available as a debian package (installable at least under Ubuntu 14.04).
Since that it is pretty easy to build a docker image such that MoleCuilder can be run inside a docker container. A Dockerfile is provided in the same blog post.
To make it even easier, I created a public repository at Docker, where the image can be pulled from directly: frederikheber:molecuilder/latest
In other words and assuming you have a working docker installation at hand, all you need to do to try the latest version is the following.
docker pull frederikheber/molecuilder:latest docker run -it frederikheber/molecuilder:latest
This will open a root shell in the container. Type
molecuilder --version
And you'll see the version installed.
For computations you need both molecuilder_server and at least one instance of molecuilder_poolworker running. The Poolworker is the number cruncher. It's a modified version of MPQC. The Server accepts computation jobs from the MoleCuilder program and pushes them on to an idle worker. We start either one in the background, logging to a file:
molecuilder_server --controllerport 10024 --workerport 10025 2&>server.log & molecuilder_poolworker --server 127.0.0.1:10025 --listen 10026 --hostname 127.0.0.1 2&>worker.log &
The server listens to MoleCuilder on port 10024, the Worker enrolls with the Server on port 10025, and finally the Worker accepts jobs on 10026. Simply choose any ports available. You can also add more than one worker at a time using GNU's parallel, which is not installed on the container,
apt-get install parallel parallel --gnu molecuilder_poolworker --server 127.0.0.1:10025 --hostname 127.0.0.1 --listen ::: 10027 10028 &
Check the server.log file to see that indeed 3 workers are in the queue now.
Well, that's all. Would you like to calculate the energy of a methane molecule?
molecuilder \ --input methane.data \ --set-output tremolo \ --set-tremolo-atomdata "id type x=3 F=3 neighbors=4" --reset 1 \ --set-parser-parameters mpqc --parser-parameters "basis=STO-3G;theory=CLHF;" \ --add-atom C --domain-position "10,10,10" \ --select-atom-by-element C --saturate-atoms \ --select-all-atoms --optimize-structure --steps 10 --deltat 1. --server-address 127.0.0.1 --server-port 10024
This should bring the largest remaining force components to about 1e-5 a.u..
Don't be overwhelmed by the lot of typing and let's explain step by step:
- First, we specify an input file (which is empty but will contain the methane on exit).
- Next, we set the output format, namely tremolo.
- And afterwards, we need to specify the tremolo format to contain the id of the atom, its element, three spatial and three gradient coordinates and at most four (covalent) neighbor ids.
- The ab-initio calculations will be done at Closed-Shell Hartree Fock (CLHF) with a simple STO-3G basis set. As we use MPQC as number cruncher, we have to give these information to the parser mpqc.
- Then, finally, we create the methane molecule by adding a carbon atom and saturating it.
- Last, all atoms are selected and the structural optimization is performed for 10 steps with an initial step length of 1 a.u.. Notice that we give the port 10024, which we told the Server to use for listening to molecuilder's jobs.
Wanna take it from here? Increase the optimization steps. Choose another basis set, e.g. 6-31G (any of MPQC's sets is fine), or Moeller-Plesset Perturbation theory 2nd order (MBPT2), ...
Some pro tips:
- have one executable running per container, i.e. start you server as
docker run --name "MoleCuilderServer" -p 10024:10024 -p 10025:10025 frederikheber/molecuilder:latest \ /usr/bin/molecuilder_server --signal 2 15 --controllerport 10024 --workerport 10025
- similarly, start you workers each in a single container, too, where we automatically extract the servers IP address
SERVERIP=`docker inspect MolecuilderServer | grep IPAddress | tail -n 1 | awk -F":" '{print $2}' | tr -d \"\ ,` docker run --name "MoleCuilderWorker" -p 10026:10026 frederikheber/molecuilder:latest \ mpirun -np 1 /usr/bin/molecuilder_poolworker --signal 2 15 --server ${SERVERIP}:10025 --listen 10026 - use GNU's parallel to start e.g. 8 workers at once, assume the above is contained in script start_worker.sh
parallel --gnu --delay 1 --ctrlc ./start_worker.sh ::: `seq 1 8`
- you can also set up docker to start containers automatically at startup, see here. But this seems to be changing in the docker API.
- install molecuilder on you local machine and use molecuildergui for the methane optimization example above.
Version v1.6.0 integrates all other required packages except Qt
To allow for an easier installation of the debian package, MoleCuilder now integrates levmar, vmg, JobMarket, CodePatterns, LinearAlgebra, and MPQC as ThirdParty packages into its distributable tarball, i.e. these dependencies no longer need to be installed extra. The only extra dependence is Qt and Qt3D (the latter being unofficially provided under DownloadRedistribution).
This makes it easy to place MoleCuilder into e.g. a Docker container and have its molecuilder_poolworker sweat away on all those fragment calculations up in some cloud, see the attached Dockerfile. Moreover, JobMarket is now fully integrated, meaning that a distinct molecuilder_server process takes care of a host of poolworkers that can munch away on the calculations in parallel. These calculations naturally are provided by molecuilder acting as the controller in this server-worker-controller implementation.
Furthermore, the fitting of empirical potential has been significantly enhanced by combining each potential fit function (e.g. dihedral angle bond) with a distinct bond model. This allows to precisely match the bond model with suitable fragments and therefore obtain more general fits of the empirical potential parametrizations. In the upcoming version, this will also work with partial charges and then we will provide some examples.
Version v1.5.4 fixes saturating bonds, fragmentation on subsets of atoms, and fitting of partial charges
We added some more complex code for saturating atoms properly. It takes fully into account already present bonds and tries to match to internally stored SphericalPointDistributions as best as possible. This gives really good initial results, i.e. building up molecules just by adding a few carbon, nitrogens and oxygens, when subsequent structure optimization relaxes everything to its proper equilibrium positions.
Moreover, we fixed the fragmentation working on just a subset of atoms, i.e. leaving out saturation water.
In the GUI, the Fragment list can now be properly sorted (and still clicked to get the fragment as set of selected atoms).
FragmentationAutomation will actually fail when the Server is unreachable and not spin forever.
ParseTremoloPotentials is gone, as it is superseded by ParseParticleParameters which is also responsible for parsing partial charges. See also SaveParticleParameters.
Note that for versions in between this and v1.5.0 we refrained from producing debian packages as these did not really bring any good change for the user. Starting from this version, it is again worth to update ... especially with what's coming up!
Version v1.5.3 brings clean QtGui<->MoleCuilder
1.5.3? Where is 1.5.1 and 1.5.2?!
To make things clear from the start, it took quite some effort to produce a clean interface between the GUI and the rest of molecuilder. Hence, v1.5.1 and v1.5.2 are sort of versions in between.
v1.5.3 basically brings mostly internal changes in form of the QtObservedInstanceBoard, namely all of the information about atoms, molecules, ... required for visual representation are copied and updated as needed. This is to make sure that things just live long enough for the sloppy GUI.
Right now, 1% of all test cases do not work in case of using the GUI as promised with v1.5.0.
Apart from that we have:
- The FitParticleCharge action is renamed to FitPartialCharge.
- long range forces working when using vmg.
- interdistance option when using Fragmentation action allows now to combine fragments up to a given order.
- electronic charges are additionally smeared for long-range electrostatic calculations which improves accuracy.
v1.5.0 brings debian packages and stable GUI
It is done!
From this version onward we provide debian packages (Ubuntu 12.04) for convenience to the user. Please note that MoleCuilder depends on several other packages, three of which had to be manually compiled and are made available at DownloadRedistribution. Especially, Qt3D was quite hard to get to work.
Furthermore, the GUI is now thread-safe!
Qt works with threads, MoleCuilder so far did not. Sure this had to clash. Now, the interface between MoleCuilder and the GUI done with Qt has been secured with mutexes and clean structures and the GUI is safe for use. All the more because it is contained in the debian package. Have fun fooling around with it.
Note that there still might be some issues. True stable execution will probably come up with v1.5.1 but early-adopters are welcome to try it out. For the moment 1 percent of all guicheck testsuite tests (see last versions about that) are still allowed to fail.
Version v1.4.12 is only a temporary step towards gui testsuite compliance
We need to switch from Codepatterns v1.2.9 to CodePatterns v1.3.0. Most of the GUI tests are failing because of race conditions in the code. CodePatterns v1.3.0 brings the Observer/Observable up to multi-threaded speed by having mutexes in place.
Hence, in this version nothing is really changed but we only switch underlying library versions on a version update.
Prepare for v1.5.0 because then the GUI tests will be working.
Version v1.4.11 features saturating atoms and an initial GUI testsuite
This is a smaller update that will some time later make a difference.
First of all, a new action has been added that allows saturating any present atom, i.e. creating methane is as simple as adding a carbon atom and saturating it.
Second, we finally have something like a testsuite for the GUI which should bring this interface up to a usable level real soon. The tests are mostly failing but this should soon be corrected.
Last, some GUI stuff has been fixed and enhanced such as the info boxes that now reveal a bit more about the atom or molecule the mouse pointer is hovering over.
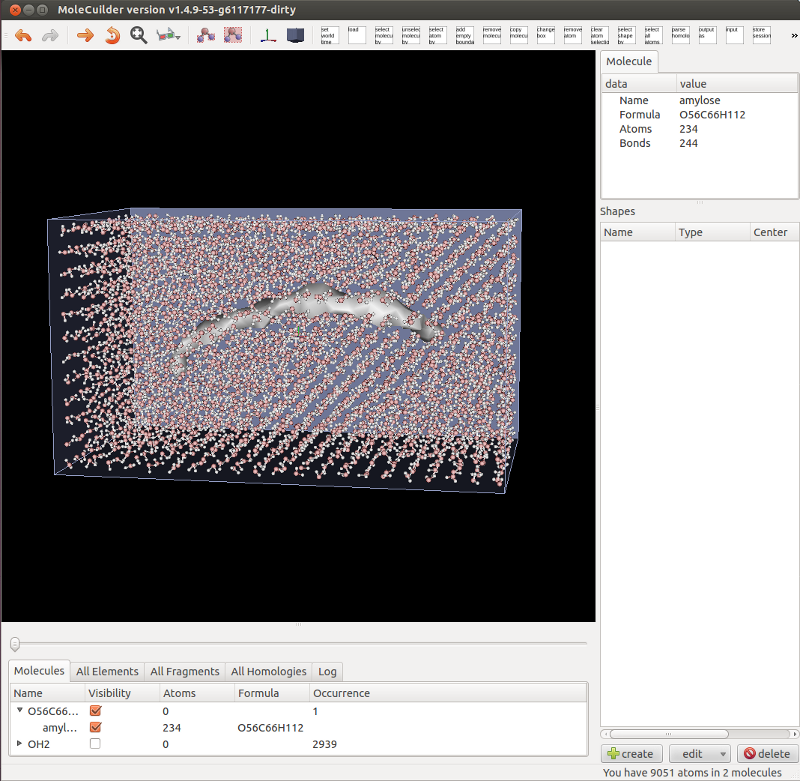
Version 1.4.10 features tesselated surfaces as new molecule view in GUI, working List of Molecules, and some more.
First of all, you can now simply flip a switch and molecules are no longer shown via their atoms and bonds but as a tesselated surface, see the image below of an amylose molecule filled with a regular grid of water molecules. This is especially useful with very large molecules (think e.g. a Tobacco Mosaic Virus or simply some large biomolecule) because a lot less triangles have to be drawn on the screen. Molecules can still be highlighted and selected by moving over or clicking onto the tesselated surface.
Notice also in the picture that the list of molecules is fully working (and hopefully) perfectly. Molecules are aggregated by their formula and can be individually selected or their visibility changed as well as for all of the same formula at once. Changing a molecule by adding an atom immediately changes the list. This rewrite became necessary as empty molecules are now automatically removed.
Furthermore, there's a number of smaller changes that should make the GUI use more easy going:
- the 3D view can now be changed via the keyboard (up/down, left/right, page-up/page-down and zooming with +/-)
- New RemoveMolecule action for convenience (before you had to select-molecules-atoms and remove-atoms)
- Selections can be pushed and popped which is useful inside MakroActions.
- Bonds are restored when undoing a remove action
- Failing actions no longer clean the queue and thus the destroy the stored session file
- when checking the command-line options for a specific action,
.../molecuilder --help fragment-molecule
is enough, actionname is no longer needed.
Version 1.4.9 features convex envelopes/tesselations, enhanced atom trajectories, complete userguide
With this version we safeguard that the userguide contains at least one example for each and every Action.
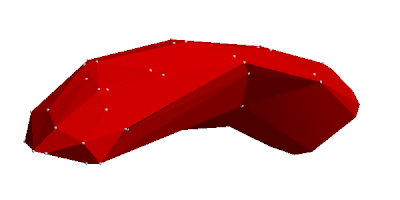
The greatest change is however that ... how to put it ... convexification of tesselated envelopes is finally working. This means that volumes of envelopes and thus the space a molecule occupies can be calculated. This is because a convex envelope's volume can be easily calculated via an inner point and general tetrahedrons. If the volume changes can be traced when transforming a non-convex envelope into a convex one, then we also know the volume of the original non-convex envelope. The volume changes however consist of either removing points ("filling up craters") or adding general tetrahedrons, both volume changes are calculable.
As an example, we give two pictures to illustrate these envelopes, here for the amylose molecule, also known as potato starch.
There is a video among the files attached to this blog post where the progress is shown and the above description of removing points and adding tetrahedrons should become a little bit clearer.
This is a very important change because knowing the volume a molecule takes up is essential in calculating densities and thus knowing how many molecules to fill into a domain. See the upcoming amylose example.
Last but not least trajectories stored in the atom class has been changed. Now, we store them as maps, not as vectors. This means that steps in the trajectory do not necessarily have to be strictly consecutive. For example, when performing a structure optimization (with debugging on) the extra trajectory steps generated for a subset of atoms are no problem anymore.
Version 1.4.8 features (almost) full valgrind validity and QtGui is more useful
Starting with version 1.4.8 we check for each version that valgrind runs through without glitches (except for some python sessions where valgrind shows too many python errors).
However, greater impact is that the GUI has been made more useful with a lot of smaller changes:
- No more clicking "ok" buttons when the dialog does not contain any queries.
- bonds are now properly updated in the 3D view.
- tooltips are shown for every query in the dialog, explaining its purpose.
- homology tab is working.
- file dialogs finally know something about the file types they are looking for.
Version 1.4.7 features enhanced hydrogen saturation procedure, Ubuntu 14.04, and structure optimization
With v1.4.7 a better hydrogen saturation procedure was implemented.
So far, we dealt with each bond to be saturated one after the other and could handle up to bond degree of three. The problem with the old code was that placement of saturation hydrogen was independent of that for another bond. This could result in hydrogens being placed on top of each other or at least very close. This occurred for example with the SLES molecule when the sulfur bonds were saturated due to its high valency.
Now, we calculate optimal places for saturation globally per molecule prior to the fragmentation procedure. After fragments have been determined and cut bonds need to be saturated, the required positions of the hydrogen are looked up from a table created by this prior calculation. This ensures that hydrogen placed by the saturation of two different bonds are optimally far away from each other.
Note that hydrogen saturation is a perturbation of the underlying Fock matrix (in the case a Hartree-Fock solver is used). Hence, resulting energies are affected and will now be slightly different.
There is a new make target extracheck that runs when mpqc (http://www.mpqc.org/) is installed and can found in the path: It compares resulting values for some molecules against stored ones. There, relative differences (for bond order of 3) of up to 1e-5 are admissable!
Furthermore, the version now compiles on Ubuntu 14.04 systems. Some changes were necessary due to new boost and gcc versions.
Last but not least a simple Structural Optimization procedure based on the fragmentation has been implemented. This is so far just using calculated forces and some fixed stepwidth dampening scheme to look for the minimum. Enhancements are coming up but will take some more time.
Version 1.4.6 features fully automated fragmentation (without JobMarket)
With v1.4.6 the src/Actions/FragmentationAction/FragmentationAutomationAction.cpp can be used even without having [doxygen:install.html JobMarket]. Fragment Jobs (i.e. each fragment is stored as an MPQC file, MPQC is executed on it and the resulting energies and forces are extracted from its output) are launched serially.
Hint: You might be tempted to specify fragment-executable as mpirun -np 6 mpqc in order to have at least multiple threads working on one fragment at a time. However, this will fail cause the option's value is checked whether it exists as a file (and there is no file called "mpirun -np 6 mpqc"). One way out is to create a small script runmpqc.sh as follows
#!/bin/sh mpirun -np 6 mpqc $@
Make it executable and use it as fragment-executable.
Another feature is that the console log is now shown inside the GUI (there is an extra widget called Log). You can click on an atom's name, e.g. C09, in the text and the atom will be (un)selected to highlight it. This comes in handy when something does not work as expected and you want to check what the code does while having the molecular system in front of you.
Last but not least: Each Action should give STATUS messages about success or failure, visible especially in the GUI (the bar at the bottom of the screen is the status bar).

 rss
rss